Research on Methods for Identifying the Tumor-specific DNA Methylation Sites in Peripheral Blood
Abstract
The recognition of tumor-specific DNA methylation sites in peripheral blood is an important research direction and hotspot in the field of liquid biopsy for cancers. Facing the requirements of liquid biopsy for cancers, this thesis makes the researches on methods for identifying tumor-specific DNA methylation sites in peripheral blood. The main results of the thesis are as follows:
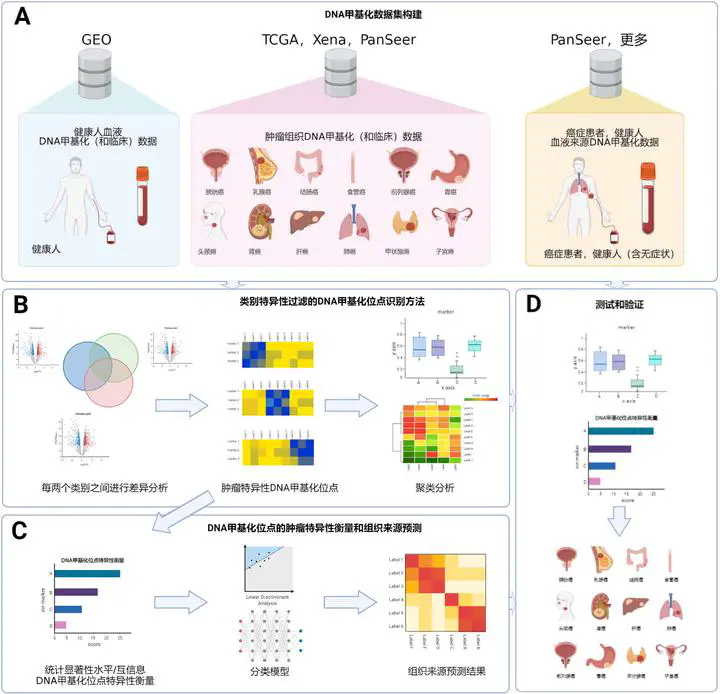
(1). This thesis integrates DNA methylation data of multiple types of tumors and blood from some tumor-related databases such as TCGA, Xena, GEO, etc., and constructs a large-scale DNA methylation dataset that can be used for identifying tumor-specific DNA methylation sites and predicting the tissue-of-origin.
(2). Considering the characteristic that the number of samples in the DNA methylation data set is much smaller than that of features, this thesis proposes a method based on category-specific filtering for identifying DNA methylation sites. Based on this method, tumor-specific DNA methylation sites for each type of tumor can be identified through differential methylation analysis between every two classes.
(3). Focusing on the problem that it is hard to screen enough tumor-specific DNA methylation sites with the increase of tumor categories, this thesis proposes a measurement method based on statistical ignificance and mutual information for measuring DNA methylation sites. And supervised classification models are used to evaluate the performance of the prediction of tumor tissue-of-origin.
The proposed method can use less information on tumor-specific DNA methylation sites to obtain better prediction performance of tumor tissue-of-origin. The effectiveness of the method for identifying tumor-specific DNA methylation sites in peripheral blood is validated on multiple data sets.