Ligandformer: A Graph Neural Network for Predicting Compound Property with Robust Interpretation
Abstract
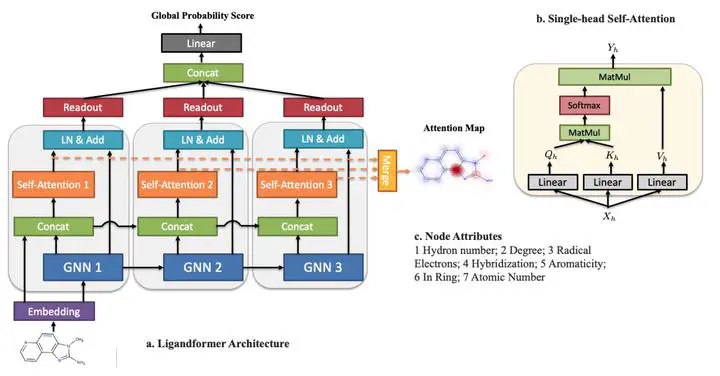
Robust and efficient interpretation of QSAR methods is quite useful to validate AI prediction rationales with subjective opinion (chemist or biologist expertise), understand sophisticated chemical or biological process mechanisms, and provide heuristic ideas for structure optimization in pharmaceutical industry. For this purpose, we construct a multi-layer self-attention based Graph Neural Network framework, namely Ligandformer, for predicting compound property with interpretation. Ligandformer integrates attention maps on compound structure from different network blocks. The integrated attention map reflects the machine’s local interest on compound structure, and indicates the relationship between predicted compound property and its structure. This work mainly contributes to three aspects: 1. Ligandformer directly opens the black-box of deep learning methods, providing local prediction rationales on chemical structures. 2. Ligandformer gives robust prediction in different experimental rounds, overcoming the ubiquitous prediction instability of deep learning methods. 3. Ligandformer can be generalized to predict different chemical or biological properties with high performance. Furthermore, Ligandformer can simultaneously output specific property score and visible attention map on structure, which can support researchers to investigate chemical or biological property and optimize structure efficiently. Our framework outperforms over counterparts in terms of accuracy, robustness and generalization, and can be applied in complex system study.
Add the publication’s full text or supplementary notes here. You can use rich formatting such as including code, math, and images.